deltametrics.plan.MorphologicalPlanform¶
- class deltametrics.plan.MorphologicalPlanform(*args, **kwargs)¶
Planform for handling the morphological method.
This Planform (called MP for short) is a wrapper/handler for the input and output from the
morphological_closing_method(). The MP is a convenient way to manage extraction of a shoreline or a delta topset area.Moreoever, the MP can be used as the input for many types of Mask objects, so it is often computationally advantageous to compute this Planform once, and then use it to create many different types of masks.
The MP provides an alternative approach to shoreline and topset area delineation to the OAM method. Depending on the input parameters chosen, this method can be faster than the OAM method, however unlike the OAM method, the accuracy and quality of the extracted planform is sensitive to the parameter values and scales inherent in the supplied inputs.
Note
It is recommended to try several parameters using a sample slice of data before computing the MP for an entire dataset, as choice of input parameters will affect the speed and quality of results!
(
Source code,png,hires.png)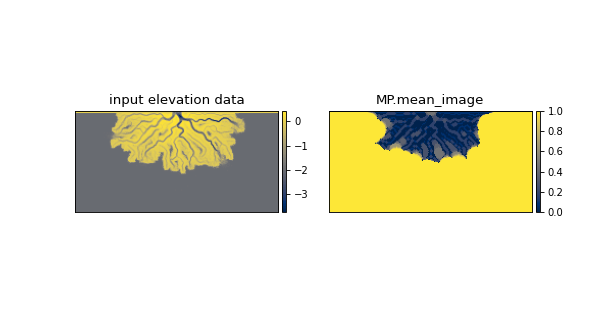
- __init__(*args, **kwargs)¶
Initialize the MorphologicalPlanform (MP).
Initializing the MP requires at least a binary input mask representing the elevation or land area of the system. A secondary input setting the maximum disk size for morphological operations can be provided.
Warning
At this time two arguments are needed! Connections between the planform object and the cube are not yet implemented.
- Parameters:
*args –
The first argument is expected to be an elevation mask, or an array which represents an elevation mask or land area. The expectation is that this input is the binary representation of the area from which you wish to identify the MP.
The second argument is the maximum disk size for morphological operations in pixels. If a cube is connected and this argument is not supplied, the inlet width will be pulled from the cube’s metadata and used to set this parameter.
**kwargs – Current supported key-word argument is ‘allow_empty’ which is a boolean argument that if True, allows the MP to be initialized with no other arguments supplied.
note:: (..) – Supplying elevation data or nonbinary data in general as the first argument to the MP will not result in an error, however the array will be coerced to be binary when morphological operations are performed. Therefore results when inputting non-binary data may not be what you expect.
Methods
__init__(*args, **kwargs)Initialize the MorphologicalPlanform (MP).
from_elevation_data(elevation_data, ...)Create a MorphologicalPlanform from elevation data.
from_mask(UnknownMask, max_disk, **kwargs)Static method for creating a MorphologicalPlanform from a mask.
show([var, ax, title, ticks, colorbar, ...])Show the planform.
Attributes
3-D array of all binary closed arrays.
Mask for below sea level pixels.
Alias for mean_image.
The public data field.
Average of all binary closing arrays.
Planform name.
Planform shape.
- __getitem__(slc)¶
Slice the planform.
Implements basic slicing for SpecialtyPlanform by passing the slc to self.data. I.e., the returned slice is
self.data[slc].
- property all_images¶
3-D array of all binary closed arrays.
- property below_mask¶
Mask for below sea level pixels.
- property composite_array¶
Alias for mean_image.
This is the array that a contour is extracted from using some threshold value when making land and shoreline masks.
- property data¶
The public data field.
This attribute must be implemented as an alias to another attribute. The choice of field is up to the developer.
- static from_elevation_data(elevation_data, max_disk, **kwargs)¶
Create a MorphologicalPlanform from elevation data.
Creates an ElevationMask from the input elevation array that is used to create the MP.
Note
Information about keyword arguments
Important
The elevation_threshold argument is implicitly required in this method, because it is required to instantiate an
ElevationMaskfrom elevation data.- Parameters:
elevation_data (
ndarray) – The elevation data to create the ElevationMask that is in turn used to create the MorphologicalPlanform.max_disk (int) – Maximum disk size to use for the morphological operations.
Examples
>>> from deltametrics.plan import MorphologicalPlanform >>> from deltametrics.sample_data.sample_data import golf
>>> golfcube = golf()
>>> MP = MorphologicalPlanform.from_elevation_data( ... golfcube["eta"][-1, :, :], elevation_threshold=0, max_disk=3 ... )
- static from_mask(UnknownMask, max_disk, **kwargs)¶
Static method for creating a MorphologicalPlanform from a mask.
- property mean_image¶
Average of all binary closing arrays.
- property name¶
Planform name.
Helpful to differentiate multiple Planform objects.
- property shape¶
Planform shape.
- show(var=None, ax=None, title=None, ticks=False, colorbar=True, colorbar_label=False)¶
Show the planform.
Display a field of the planform, called by attribute name.
- Parameters:
var (
str) – Which field to show. Must be an attribute of the planform. show will look for another attribute describing theVariableInfofor that attribute namedself._<var>_varinfoand use that to style the plot, if found. If this VariableInfo is not found, the default is used.label (
bool, str, optional) – Display a label of the variable name on the plot. Default is False, display nothing. Iflabel=True, the label name from theVariableSetis used. Other arguments are attempted to coerce to str, and the literal is diplayed.colorbar (
bool, optional) – Whether a colorbar is appended to the axis.colorbar_label (
bool, str, optional) – Display a label of the variable name along the colorbar. Default is False, display nothing. Iflabel=True, the label name from theVariableSetis used. Other arguments are attempted to coerce to str, and the literal is diplayed.ax (
Axesobject, optional) – A matplotlib Axes object to plot the section. Optional; if not provided, a call is made toplt.gca()to get the current (or create a new) Axes object.